A ‘word processor’ for genes – Scientists discover a fundamental new mechanism for biological programming
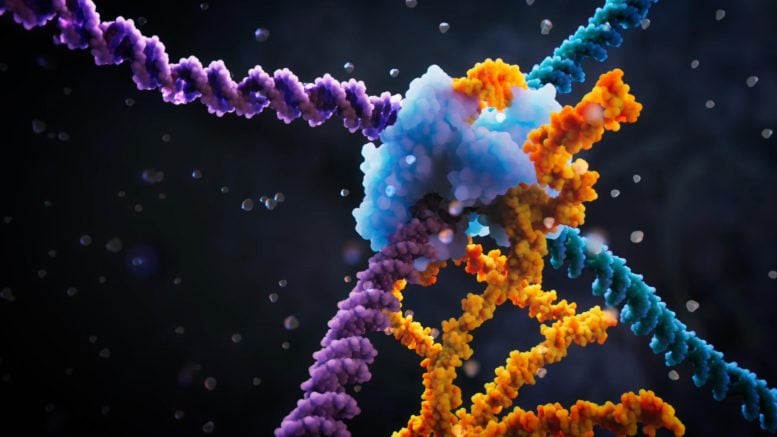
Visualization of the bridge recombinase mechanism. Credit: Visual Science
Arc Institute scientists have discovered the bridge recombinase mechanism, a revolutionary tool that enables fully programmable DNA rearrangements.
Finding them, detailed in a recent one Nature publication, is the first DNA recombinase to use a non-coding RNA for sequence-specific selection of target and donor DNA molecules. This bridge RNA is programmable, allowing the user to specify any desired genomic target sequence and any donor DNA molecule to be inserted.
The research was conducted in collaboration with the laboratories of Silvana Konermann, Arc Institute Principal Investigator and Assistant Professor of Biochemistry at Stanford University, and Hiroshi Nishimasu, Professor of Structural Biology at the University of Tokyo.
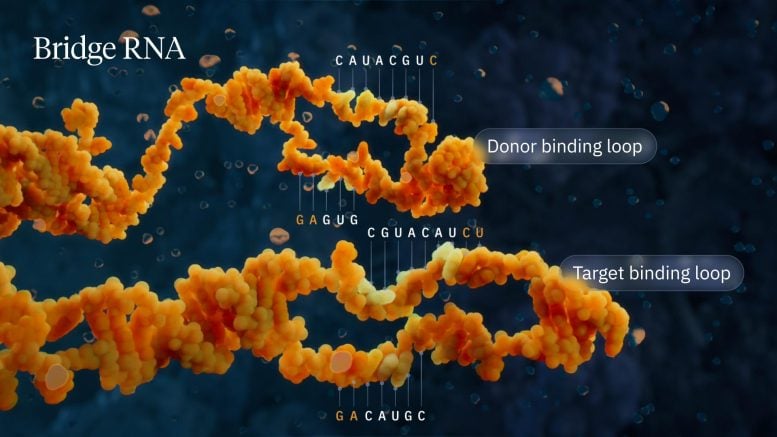
Visualization of the bridge recombinase mechanism highlighting donor and target binding loops. Credit: Visual Science
A new era of genetic programming
“The bridge RNA system is a fundamentally new mechanism for biological programming,” said Dr. Patrick Hsu, senior author of the study and an Arc Institute principal investigator and University of California, Berkeley Assistant Professor of Bioengineering. “Bridge recombination can universally modify genetic material through sequence-specific insertions, deletions, inversions, and more, enabling a word processor for the living genome beyond CRISPR.”
The bridge recombination system comes from insertion sequence 110 (IS110) elements, one of countless types of transposable elements—or “jumping genes”—that cut and paste to move within and between microbial genomes. Transposable elements are found in all life forms and have evolved into professional DNA manipulation machines to survive. IS110 elements are very minimal, consisting only of a gene encoding the recombinase enzyme, plus flanking DNA segments that have so far remained a mystery.
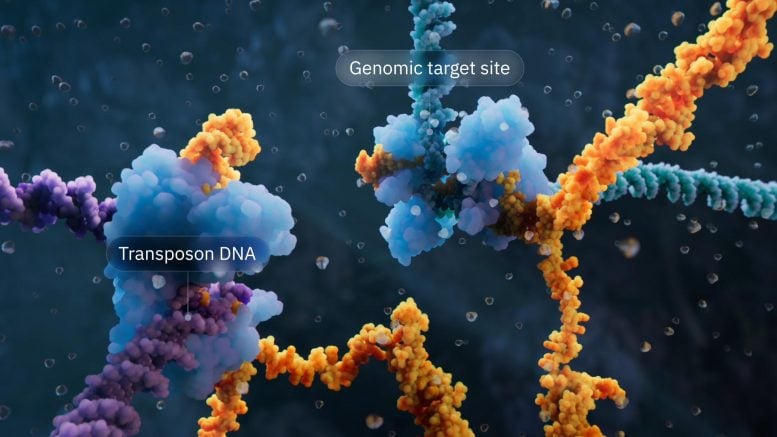
Visualization of the bridge recombinase mechanism highlighting the transposon DNA and genomic target location. Credit: Visual Science
The Advanced Mechanism of Bridge RNA
The Hsu lab discovered that when IS110 is detached from a genome, the ends of the non-coding DNA join together to produce an RNA molecule—bridge RNA—that folds into two loops. One loop binds to the IS110 element itself, while the other loop binds to the target DNA where the element will be inserted. Bridge RNA is the first example of a bispecific guide molecule, specifying the sequence of the target and donor DNA through base-pairing interactions.
A team of researchers from the Arc Institute have discovered the bridge recombinase mechanism, a precise and powerful tool to recombine and rearrange DNA in a programmable way. Going far beyond programmable genetic scissors like CRISPR, the bridging recombinase mechanism allows scientists to specify not only the target DNA to be modified, but also the donor material that needs to be recognized so that they can insert new functional genetic material, cut out faulty DNA. or reverse any two sequences of interest. Find out more in this short video visualizing key aspects of the bridge recombination mechanism. Credit: Visual Science
Each loop of the bridge RNA is independently programmable, allowing researchers to mix and match any target and donor DNA sequences of interest. This means that the system can go far beyond its natural role of inserting the IS110 element itself, instead enabling the insertion of any desired genetic cargo—such as a functional copy of a faulty, disease-causing gene—into any genomic location. In this work, the team demonstrated over 60% efficiency of inserting a desired gene into E. coli with over 94% specificity for the exact genomic location.
“These programmable bridge RNAs distinguish IS110 from other known recombinases, which lack an RNA component and cannot be programmed,” said co-lead author Nick Perry, a bioengineering graduate student at UC Berkeley. “It’s like the bridge RNA is a universal power adapter that makes the IS110 compatible with any outlet.”

Patrick Hsu, Nick Perry, and Matt Durrant discuss the newly discovered bridge recombinase mechanism. Credit: Ray Rudolph
Collaborative research and future implications
The Hsu lab’s discovery is complemented by their collaboration with Dr. Hiroshi Nishimasu at the University of Tokyo, also published on June 26 at Nature. The Nishimasu lab used cryo-electron microscopy to determine the molecular structures of the recombinase bridge RNA complex bound to target and donor DNA, sequentially progressing through the key steps of the recombination process.

Januka Athukoralage, Nicholas Perry, Silvana Konermann, Matthew Durrant, Patrick Hsu, James Pai and Aditya Jangid. Credit: Ray Rudolph
With further exploration and development, the bridge mechanism promises to usher in a third generation of RNA-directed systems, expanding beyond DNA cutting and RNAi mechanisms of CRISPR and RNA interference (RNAi) to provide a unified mechanism for programmable DNA rearrangements. Critical to the further development of the bridge recombinase system for mammalian genome design, the bridge recombinase joins both DNA strands without releasing nicked DNA fragments—bypassing a key limitation of modern editing technologies. the genome.
“The bridge recombination mechanism solves some of the most fundamental challenges faced by other genome editing methods,” said research co-leader Matthew Durrant, a senior scientist at Arc. “The ability to programmatically rearrange any two DNA molecules opens the door to advances in genome design.”
References:
“Direct RNA Programmable Recombination of Target RNA and Donor DNA” by Matthew G. Durrant, Nicholas T. Perry, James J. Pai, Aditya R. Jangid, Januka S. Athukoralage, Masahiro Hiraizumi, John P. McSpedon, April Pawluk, Hiroshi Nishimasu, Silvana Konermann, and Patrick D. Hsu, 26 June 2024, Nature.
DOI: 10.1038/s41586-024-07552-4
“Structural Mechanism of RNA-Directed Recombination” by Masahiro Hiraizumi, Nicholas T. Perry, Matthew G. Durrant, Teppei Soma, Naoto Nagahata, Sae Okazaki, Januka S. Athukoralage, Yukari Isayama, James J. Pai, Aprila Pawluk , Konermann, Keitaro Yamashita, Patrick D. Hsu, and Hiroshi Nishimasu, 26 Jun 2024, Nature.
DOI: 10.1038/s41586-024-07570-2










Post Comment